OnkoSight AdvancedTM Pan-Heme Fusion Panel
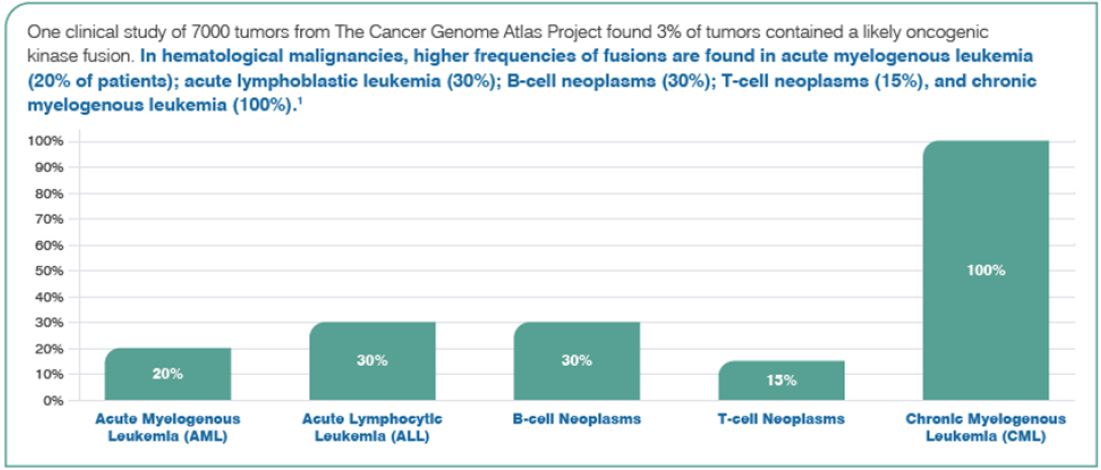
Gene fusions have become increasingly important as diagnostic and therapeutic targets in cancer. Fusions are a type of somatic alteration leading to cancer associated with up to 20% of cancer morbidity.
Fusions arise due to genomic rearrangements that include chromosomal inversion, interstitial deletions, duplications, and translocations. Conventional methodologies such as chromosome analysis, fluorescence in situ hybridization (FISH), and RT-PCR have been used to detect gene rearrangements, but these procedures have limitations. For example, t(12;21)(p13;q22) with ETV6-RUNX1 is often missed in the analysis of chromosomes. Moreover, translocations that involve KMT2A (MLL), which has >100 partner genes with numerous different breakpoints, are not fully covered by the limited number of primer sets used in RT-PCR.
Clinical Utility of Gene Fusions in Hematology
- Diagnosis and classification of various myeloid and lymphoid neoplasms
- Subgrouping and prognostication in leukemia
- Modification of treatment intensity or early stem cell transplantation (i.e., KMT2A and BCR-ABL1 translocations)
- Eligibility to targeted molecular drugs (e.g., tyrosine kinase inhibitors)
- Identification of potential clinical trial eligibility
Test Information
OnkoSight AdvancedTM Pan-Heme Fusion NGS Panel (73 genes)
TP02-2
ABL1 |
ABL2 |
ALK |
BCL11B |
BCL2 |
BCL6 |
BCR |
BIRC3 |
CBFB |
CCND1 |
CCND3 |
CDK6 |
CHD1 |
CHIC2 |
CIITA |
CREBBP |
CRLF2 |
CSF1R |
DEK |
DUSP22 |
EBF1 |
EIF4A1 |
EPOR |
ERG |
ETV6 |
FGFR1 |
GUS2 |
IKZF1 |
IKZF2 |
IKZF3 |
JAK2 |
KAT6A |
KLF2 |
KMT2A |
MALT1 |
MECOM |
MKL1 |
MLF1 |
MLLT10 |
MLLT4 |
MYC |
MYH11 |
NF1 |
NFKB2 |
NOTCH1 |
NTRK3 |
NUP214 |
NUP98 |
P2RY8 |
PAG1 |
PAX5 |
PBX1 |
PDCD1L G2 |
PDGFRA |
PDGFRB |
PICALM |
PML |
PRDM16 |
PTK2B |
RARA |
RBM15 |
ROS1 |
RUNX1 |
RUNX1T1 |
SEMA6A |
SETD2 |
STIL |
TAL1 |
TCF3 |
TFG |
TP63 |
TYK2 |
ZCCHC7 |
GenPath® also offers single-gene testing. Healthcare providers should only order panels if each test in the panel is medically necessary.
Download a sample report here.
References:
1. Nikanjam M, Okamura R, Barkauskas DA, Kurzrock R. Targeting fusions for improved outcomes in oncology treatment. Cancer. 2020 Mar 15;126(6):1315-1321. doi: 10.1002/cncr.32649. Epub 2019 Dec 3. PMID: 31794076; PMCID: PMC7050395.
2. Foltz SM, Gao Q, Yoon CJ, Sun H, Yao L, Li Y, Jayasinghe RG, Cao S, King J, Kohnen DR, Fiala MA, Ding L, Vij R. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat Commun. 2020 May 29;11(1):2666. doi: 10.1038/s41467-020-16434-y. PMID: 32471990; PMCID: PMC7260243.
3. Borahm Kim, Hyeonah Lee, Saeam Shin, Seung-Tae Lee, Jong Rak Choi, Clinical Evaluation of Massively Parallel RNA Sequencing for Detecting Recurrent Gene Fusions in Hematologic Malignancies, The Journal of Molecular Diagnostics, Volume 21, Issue 1, 2019.

